- Messages
- 2,145
- Reaction score
- 5
- Points
- 28
- Thread Starter
- #21
Re: Genetics: Unlocking Humanity's Past and Future
Genetics: Future
Scientists have managed to coax living cells into making carbon-silicon bonds, demonstrating for the first time that nature can incorporate silicon - one of the most abundant elements on Earth - into the building blocks of life.
While chemists have achieved carbon-silicon bonds before - they’re found in everything from paints and semiconductors to computer and TV screens - they’ve so far never been found in nature, and these new cells could help us understand more about the possibility of silicon-based life elsewhere in the Universe.
After oxygen, silicon is the second most abundant element in Earth’s crust, and yet it has nothing to do with biological life.
Why silicon has never been incorporated into any kind of biochemistry on Earth has been a long-standing puzzle for scientists, because, in theory, it would have been just as easy for silicon-based lifeforms to have evolved on our planet as the carbon-based ones we know and love.
Not only are carbon and silicon both extremely abundant in Earth’s crust - they’re also very similar in their chemical make-up.
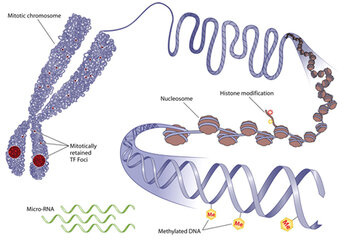
One of the most important features carbon and silicon share is the ability to form bonds with four atoms at the same time. This means they're capable of linking together the long chains of molecules needed to form the basis of life as we know it - proteins and DNA.
And yet, silicon-based lifeforms do not exist outside the Star Trek universe - as far as we know.
"No living organism is known to put silicon-carbon bonds together, even though silicon is so abundant, all around us, in rocks and all over the beach," says one of the researchers, Jennifer Kan from Caltech.
To be clear, Kan and her team played a big role in getting living cells to achieve carbon-silicon bonds - this was not something that the cell could have easily done on its own.
But the experiment is proof that these bonds can be formed in nature - so long as you have the right conditions.
The researchers started by isolating a protein that occurs naturally in the bacterium Rhodothermus marinus, which thrives in the hot springs of Iceland.
They liked this protein, called cytochrome c enzyme, because while its main role is to transport electrons through the cells, lab tests revealed that it could facilitate the kinds of bonds that could attach silicon atoms to carbon.
After isolating the protein, they inserted the gene for it into some E. coli bacteria to if it could facilitate the production of carbon-silicon bonds inside its living cells.
The first iteration of these silicon-engineered bacteria didn’t do much, but the team continued to mutate the protein gene within a specific region of the E. coli genome until something very cool happened.
"After three rounds of mutations, the protein could bond silicon to carbon 15 times more efficiently than any synthetic catalyst," Aviva Rutkin reports for New Scientist.
The fact that this bacterium has been engineered to produce carbon-silicon bonds more efficiently than chemists can in the lab is exciting for two reasons. First, it offers up a better way to produce the carbon-silicon bonds we need to make things like pharmaceuticals, agricultural chemicals, and fuels.
"This is something that people talk about, dream about, wonder about," Annaliese Franz from the University of California, Davis, who wasn’t involved in the research, told New Scientist.
"Any pharmaceutical chemist could read this on Thursday and on Friday decide they want to take this as a building block that they could potentially use."
Secondly, it signifies that a lifeform could potentially be at least partially based on silicon, and if the researchers continue to grow these kinds of bacteria, we could get a better understanding of what they could look like.
"This study shows how quickly nature can adapt to new challenges," one of the team, Frances Arnold, said in a press statement.
"The DNA-encoded catalytic machinery of the cell can rapidly learn to promote new chemical reactions when we provide new reagents and the appropriate incentive in the form of artificial selection. Nature could have done this herself if she cared to."
The research has been published in Science.
Genetics: Future
For the first time, living cells have formed carbon-silicon bonds
Life—but not as we know it.

Life—but not as we know it.
Scientists have managed to coax living cells into making carbon-silicon bonds, demonstrating for the first time that nature can incorporate silicon - one of the most abundant elements on Earth - into the building blocks of life.
While chemists have achieved carbon-silicon bonds before - they’re found in everything from paints and semiconductors to computer and TV screens - they’ve so far never been found in nature, and these new cells could help us understand more about the possibility of silicon-based life elsewhere in the Universe.
After oxygen, silicon is the second most abundant element in Earth’s crust, and yet it has nothing to do with biological life.
Why silicon has never been incorporated into any kind of biochemistry on Earth has been a long-standing puzzle for scientists, because, in theory, it would have been just as easy for silicon-based lifeforms to have evolved on our planet as the carbon-based ones we know and love.
Not only are carbon and silicon both extremely abundant in Earth’s crust - they’re also very similar in their chemical make-up.
One of the most important features carbon and silicon share is the ability to form bonds with four atoms at the same time. This means they're capable of linking together the long chains of molecules needed to form the basis of life as we know it - proteins and DNA.
And yet, silicon-based lifeforms do not exist outside the Star Trek universe - as far as we know.
"No living organism is known to put silicon-carbon bonds together, even though silicon is so abundant, all around us, in rocks and all over the beach," says one of the researchers, Jennifer Kan from Caltech.
To be clear, Kan and her team played a big role in getting living cells to achieve carbon-silicon bonds - this was not something that the cell could have easily done on its own.
But the experiment is proof that these bonds can be formed in nature - so long as you have the right conditions.
The researchers started by isolating a protein that occurs naturally in the bacterium Rhodothermus marinus, which thrives in the hot springs of Iceland.
They liked this protein, called cytochrome c enzyme, because while its main role is to transport electrons through the cells, lab tests revealed that it could facilitate the kinds of bonds that could attach silicon atoms to carbon.
After isolating the protein, they inserted the gene for it into some E. coli bacteria to if it could facilitate the production of carbon-silicon bonds inside its living cells.
The first iteration of these silicon-engineered bacteria didn’t do much, but the team continued to mutate the protein gene within a specific region of the E. coli genome until something very cool happened.
"After three rounds of mutations, the protein could bond silicon to carbon 15 times more efficiently than any synthetic catalyst," Aviva Rutkin reports for New Scientist.
The fact that this bacterium has been engineered to produce carbon-silicon bonds more efficiently than chemists can in the lab is exciting for two reasons. First, it offers up a better way to produce the carbon-silicon bonds we need to make things like pharmaceuticals, agricultural chemicals, and fuels.
"This is something that people talk about, dream about, wonder about," Annaliese Franz from the University of California, Davis, who wasn’t involved in the research, told New Scientist.
"Any pharmaceutical chemist could read this on Thursday and on Friday decide they want to take this as a building block that they could potentially use."
Secondly, it signifies that a lifeform could potentially be at least partially based on silicon, and if the researchers continue to grow these kinds of bacteria, we could get a better understanding of what they could look like.
"This study shows how quickly nature can adapt to new challenges," one of the team, Frances Arnold, said in a press statement.
"The DNA-encoded catalytic machinery of the cell can rapidly learn to promote new chemical reactions when we provide new reagents and the appropriate incentive in the form of artificial selection. Nature could have done this herself if she cared to."
The research has been published in Science.
Attachments
Last edited: