- Messages
- 2,145
- Reaction score
- 5
- Points
- 28
GENETICS: WE ARE ALL AFRICANS
by George Busby |October 31, 2016
Three new studies reveal the diversity of early human populations and help pin down when we left Africa.
SUMMARYby George Busby |October 31, 2016
Three new studies reveal the diversity of early human populations and help pin down when we left Africa.
- All humans across the globe right now—no exception—trace their roots to Africa.
[*]Tracing the ancestor of all humans on Earth is a direct consequence of the Human Genome Project (HGP), the international scientific research project with the goal of determining the sequence of nucleotide base pairs which make up human DNA, and of identifying and mapping all of the genes of the human genome from both a physical and a functional standpoint.
[*]Genetic and paleontological evidence show humans only started to leave Africa between 60,000 and 70,000 years ago.
[*]What set this in motion is a major cataclysm, something to do with major climatic shifts that were happening around that time—a sudden cooling in the Earth’s climate driven by the onset of one of the worst parts of the last Ice Age. - The last of our African ancestors who managed to survive and escape Africa were reduced to a population as low as 10,000—humanity was holding on by a thread at this time. It's the ultimate drama of human existence, one to stand for ages to come.
[*]Genetics will revolutionize human societies in many ways not easy to imagine right now.
Humans are a success story like no other. We are now living in the “Anthropocene” age, meaning much of what we see around us has been made or influenced by people. Amazingly, all humans alive today – from the inhabitants of Tierra del Fuego on the southern tip of the Americas to the Sherpa in the Himalayas and the mountain tribes of Papua New Guinea – came from one common ancestor.
We know that our lineage arose in Africa and quickly spread to the four corners of the globe. But the details are murky. Was there just one population of early humans in Africa at the time? When exactly did we first leave the continent and was there just one exodus? Some scientists believe that all non-Africans today can trace their ancestry back to a single migrant population, while others argue that there were several different waves of migration out of Africa.
Now, three new studies mapping the genetic profiles of more than 200 populations across the world, published in Nature, have started to answer some of these questions.
Image via Preben Hjort, Mayday Film.
Out of Africa
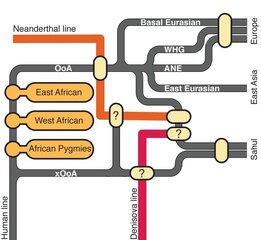
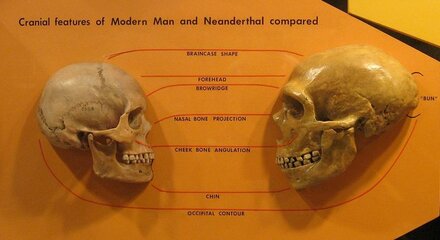
Humans initially spread out of Africa through the Middle East, ranging further north into Europe, east across Asia and south to Australasia. Later, they eventually spread north-east over the top of Beringia into the Americas. We are now almost certain that on their way across the globe, our ancestors interbred with at least two archaic human species, the Neanderthals in Eurasia, and the Denisovans in Asia.
Genetics has been invaluable in understanding this past. While hominin fossils hinted that Africa was the birthplace of humanity, it was genetics that proved this to be so. Patterns of genetic variation – how similar or different people’s DNA sequences are – have not only shown that most of the diversity we see in humans today is present within Africa, but also that there are fewer differences within populations the further you get from Africa.
These observations support the “Out of Africa” model; the idea that a small number of Africans moved out of the continent – taking a much reduced gene-pool with them. This genetic bottleneck, and the subsequent growth of non-African populations, meant that there was less genetic diversity to go round, and so there are fewer differences, on average, between the genomes of non-Africans compared to Africans.
When we scan two genomes to identify where these differences, or mutations, lie, we can estimate how long in the past those genomes split from each other. If two genomes share long stretches with no differences, it’s likely that their common ancestor was in the more recent past than the ancestor of two genomes with shorter shared stretches. By interrogating the distribution of mutations between African and non-African genomes, two of the papers just about agree that the genetic bottleneck caused by the migration out of Africa occurred roughly 60,000 years ago. This is also broadly in line with dating from archaeological investigations.
Their research also manages to settle a long-running debate about the structure of African populations at the beginning of the migration. Was the small group of humans who left Africa representative of the whole continent at that time, or had they split off from more southerly populations earlier?
SGDP model of the relationships among diverse humans (select ancient samples are shown in red) that fits the data.
Image via Swapan Mallick, Mark Lipson and David Reich.
Image via Swapan Mallick, Mark Lipson and David Reich.
The Simons Genome Diversity Project compared the genomes of 142 worldwide populations, including 20 from across Africa. They conclusively show that modern African hunter-gatherer populations split off from the group that became non-Africans around 130,000 years ago and from West Africans around 90,000 years ago. This indicates that there was substantial substructure of populations in Africa prior to the wave of migration. A second study, led by Danish geneticist Eske Willersev, with far fewer African samples, used similar methods to show that divergence within Africa also started before the migration, around 125,000 years ago.
More migrations?
Following the move out of the continent, the pioneers must then have journeyed incredibly quickly to Australia. The Danish study, the most comprehensive analysis of Aboriginal Australian and Papuan genomes to date, is the first to really examine the position of Australia at the end of the migration.
They found that the ancestors of populations from “Sahul” – Tasmania, Australia and New Guinea – split from the common ancestor of Europeans and Asians 51,000-72,000 years ago. This is prior to their split from each other around 29,000-55,000 years ago, and almost immediately after the move out of Africa. This implies that the group of people who ended up in the Sahul split away from others almost as soon as the initial group left Africa. Substantial mixing with Denisovans is only seen in Sahulians, which is consistent with this early split.
Crucially, because the ancestors of modern-day Europeans and Asians hadn’t split in two at this point, we think that they must have still been somewhere in western Eurasia at this point. This means that there must have been a second migration from west Eurasia into east Asia later on. The Simons Genome Diversity Project study, by contrast, albeit with a far smaller sample of Sahulian genomes, found no evidence for such an early Sahulian split. It instead shows that the ancestors of East Asians and Sahulians split from western Eurasians before they split from each other, and therefore that Denisovan admixture occurred after the former split from each other.
A graphic representation of the interaction between modern and archaic human lines, showing traces of an early out of Africa (xOoA) expansion within the genome of modern Sahul populations. Image via Dr. Mait Metspalu, Estonian Biocentre
.Meanwhile, a third paper proposes an earlier, “extra” migration out of Africa, some 120,000 years ago. This migration is only visible in the genomes of a separate set of Sahulians sequenced as part of the Estonian Biocentre Human Genome Diversity Panel. Only around 2% per cent of these genomes can be traced to this earlier migration event, which implies that this wave can’t have many ancestors left in the present day. If true (the two other papers find little support for it), this suggests that there must have been a migration across Asia prior to the big one about 60,000 years ago, and that anatomically modern human populations left Africa earlier than many think.
Whatever the reality of the detail of the Out of Africa event, these studies provide some benchmarks for the timings of some of the key events. Importantly, they are also a huge resource of over 600 new and diverse human genomes that provide the genomics community with the opportunity for further understanding of the paths our ancestors took towards the Anthropocene.
George Busby, Research Associate in Statistical Genomics, University of Oxford
This article was originally published on The Conversation. Read the original article.
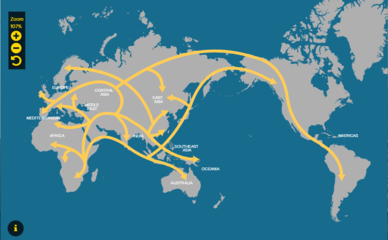
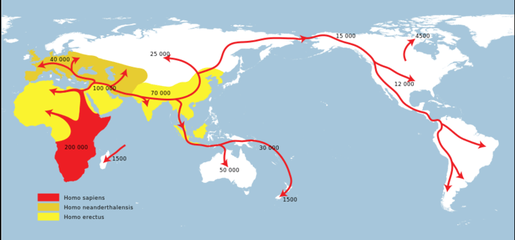
MIGRATION ROUTES

When humans first ventured out of Africa some 60,000 years ago, they left genetic footprints still visible today. By mapping the appearance and frequency of genetic markers in modern peoples, we create a picture of when and where ancient humans moved around the world. These great migrations eventually led the descendants of a small group of Africans to occupy even the farthest reaches of the Earth.
Our species is an African one: Africa is where we first evolved, and where we have spent the majority of our time on Earth. The earliest fossils of recognizably modern Homo sapiens appear in the fossil record at Omo Kibish in Ethiopia, around 200,000 years ago. Although earlier fossils may be found over the coming years, this is our best understanding of when and approximately where we originated.
According to the genetic and paleontological record, we only started to leave Africa between 60,000 and 70,000 years ago. What set this in motion is a major cataclysm, something to do with major climatic shifts that were happening around that time—a sudden cooling in the Earth’s climate driven by the onset of one of the worst parts of the last Ice Age. This cold snap would have made life difficult for our African ancestors, and the genetic evidence points to a sharp reduction in population size around this time. In fact, the human population likely dropped to fewer than 10,000. We were holding on by a thread.
Once the climate started to improve, after 70,000 years ago, we came back from this near-extinction event. The population expanded, and some intrepid explorers ventured beyond Africa. The earliest people to colonize the Eurasian landmass likely did so across the Bab-al-Mandab Strait separating present-day Yemen from Djibouti. These early beachcombers expanded rapidly along the coast to India, and reached Southeast Asia and Australia by 50,000 years ago. The first great foray of our species beyond Africa had led us all the way across the globe.
Slightly later, a little after 50,000 years ago, a second group appears to have set out on an inland trek, leaving behind the certainties of life in the tropics to head out into the Middle East and southern Central Asia. From these base camps, they were poised to colonize the northern latitudes of Asia, Europe, and beyond.
Around 20,000 years ago a small group of these Asian hunters headed into the face of the storm, entering the East Asian Arctic during the Last Glacial Maximum. At this time the great ice sheets covering the far north had literally sucked up much of the Earth’s moisture in their vast expanses of white wasteland, dropping sea levels by more than 300 feet. This exposed a land bridge that connected the Old World to the New, joining Asia to the Americas. In crossing it, the hunters had made the final great leap of the human journey. By 15,000 years ago they had penetrated the land south of the ice, and within 1,000 years they had made it all the way to the tip of South America. Some may have even made the journey by sea.
The story doesn’t end there, of course. The rise of agriculture around 10,000 years ago—and the population explosion it created—has left a dramatic impact on the human gene pool. The rise of empires, the astounding oceangoing voyages of the Polynesians, even the extraordinary increase in global migration over the past 500 years could all leave traces in our DNA. There are many human journey questions waiting to be asked and answered.
What stories are waiting to be told in your own DNA?
Attachments
Last edited: